Impact of CpG site specific methylation on silencing protein expression of GSTP1 in breast cancer
Valla Fongchaiya, BSc *, Pichet Sampatanukul, MD, MSc*, and Kris Chatamra, MD, FRCS **
*Department of Pathology, Faculty of Medicine, Chulalongkorn University and
**Queen Sirikit Center for Breast Cancer, King Chulalongkorn Memorial Hospital, Thai Red Cross Society.
Correspondence: Valla Fongchaiya, BSc.,
Department of Pathology, Faculty of Medicine, Chulalongkorn University.
Email address: Valla.fongchaiya@gmail.com
Received 26 April 2013; Accepted 27 May 2013.
ABSTRACT
Introduction: From previous study with Methylation specific polymerase chain reaction (MSP), there were cases of DNA methylation showing positive protein expression. Some CpG sites of the primer were speculated to have no effect on protein expression.
Methods: Forty-six cases were selected including the two problem cases from our previous study samples of 88. The DNA extracts were sequenced after bisulfite conversion for the epigenetic part of the GSTP1 gene, 298 base pairs, which comprised 38 CpG sites. Methylation status at the sites 10-13 and 20-22 representing in the primer used in MSP study was correlated with the expression of protein to detect the active and inactive sites. result: The two problem cases revealed methylation of CpG sites no.11, 12, 13 and no. 21, 22, suggested that these were inactive sites. The sites no. 10 and 20 were regarded as active sites. The validity was confirmed on the methylation data of the rest of 44 cases. There were 11 cases showing methylated cytosine at either CpG site no. 10 or 20. All cases showed negative for the protein. On the other hand, of the 21 cases with positive protein, none elicited methylation at CpG sites no.10 and 20.
Conclusion: The impact of location of CpG sites on the silencing of the protein expression of GSTP1 is present. There are at least two active sites at no.10 and 20. Further studies to verify the other active sites are encouraging.
Keywords: DNA methylation, CpG site, Protein expression, GSTP1, Breast cancer
Introduction
DNA methylation is a well-established epigenetic mechanism in regulation of protein expression1. The conjoint mechanism is unclear whether the inhibition is dependent on level of methylation or site specific of CpG that is methylated 2 3. Regarding our finding in the previous study, there were some cases of hypermethylation found and protein present for GSTP1 in invasive breast carcinoma3. We speculated that this event might be due to positive methylation in “inactive” CpG sites. To prove this concept of some sites when methylated will affect the gene silencing on encoding protein while other sites when methylated will not, we carried out the study with Bisulfite sequencing technique to reveal the methylation status of the CpG sites that were comprised in the primer of former MSP study and verify the concept statement.
MATERIAL AND METHOD
Sample collection
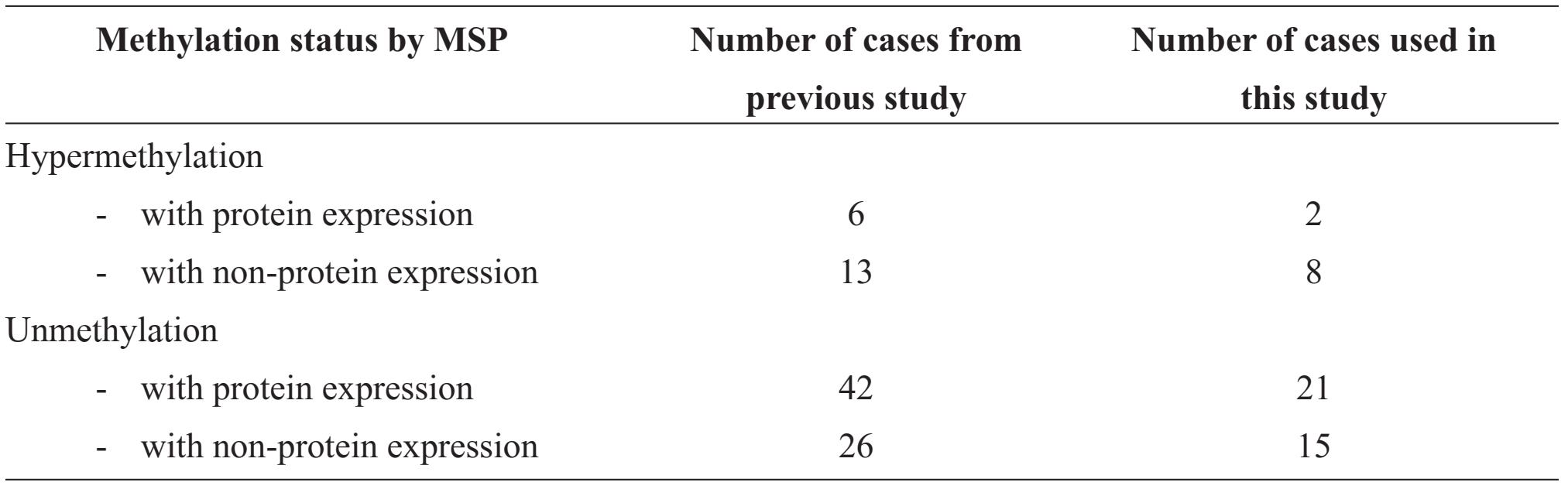
From our previous study, the correlation between hypermethylation by Methylation specific PCR (MSP) technique and protein expression by immunohistochemistry were categorized into four groups of samples (shown in table1). This studied samples were randomly drawn from each group with an approximate ratio; they comprised 2 cases of hypermethylation with protein expression, 8 cases of hypermethylation with non-protein expression, 21 cases of unmethylation with protein expression and 15 cases of unmethylation with non-protein expression.
DNA isolation
Fresh tumor tissues of breast cancer kept in the refrigerator (-80°C) were thawed. The source and specimen data had mentioned in the previous paper. DNA was isolated by phenol-chloroform method; in brief, incubate the tissue samples with proteinase K for 16-18 hours before phenol extraction and sodium acetate-isopropanol precipitation. Concentration of DNA was verified by Nanodrop 2000c Thermo Scientific.
Bisulfite sequencing
- Bisulfite reaction
Genomic DNA 400 ng was performed using EZ DNA Methylation-Gold kit™ (Zymo Research). CT conversion reagent consisted of 900 pl distill water, 300 pl M-dilution buffer and 50 pl M-dissolving buffer. Each reaction used 130 pl of the CT conversion reagent to 20 pl of DNA sample in a PCR tube. The tubes were placed in Thermal Cycler, incubated at 98 °C for 10 minutes and 64 °C for 2.5 hours. Products could be stored at 4°C. More details are elaborated elsewhere3.
- PCR reaction
PCR was performed in Vertiri 96 well thermo cycler Applied Biosystem™ for 45 cycles. The PCR program started with activation of the polymerase at 95 °C for 15 min followed by dena-turation at 95 °C for 1 min, annealing at 64 °C for 1 min and extension at 72 °C for 1 min followed by a final 4 mins extension at 72 °C and cooling at 4°C.
The reaction was assembled in a final volume at 30 pl, containing 0.6 pl of dNTP, 3 pl of 10x buffer, 0.3 pl of primer, 2 pl of bisulfite treated DNA and 21.1 μl of water.
Table 1 Number of cases in the strata of methylation and protein expression of GSTP1 in breast cancer of the previous and present studies
PCR products were run on 8% polyacrylamide gel to verify the correct size.
Before the sequencing reaction, PCR products were purified with ExoSAP™ for cleanup of PCR products that eliminates unincorporated primers and dNTPs.
- Sequencing reaction
The sequencing reaction contains DNA template from purified PCR product, reverse primer 0.8 pmol/pl, sequencing buffer and dNTP/ ddNTP substrate from the BigDye terminator v3.1 Cycle Sequencing kit (Applied Biosystem, USA). The mixture was performed on Thermocycler and program started with 95 °C for 1 minute and 96 °C for 10 sec, 50°C for 5 sec, 60°C for 4 minutes for 25 cycles and 60 °C for 4 minutes in the final step.
DNA bisulfite sequencing was analyzed by automate capillary electrophoresis and the reaction was purified by 3M sodium acetate precipitation before capillary electrophoresis using Genetic analyzer ABI3130.
Immunohistochemistry and Methylation specific PCR
Data were obtained from the previous study and the method descriptions were referred to the paper3. In order to verify the result of immunohistochemistry, the relevant stained slides were reviewed, scanned with Image Scope®, and 400 tumor cells were counted for the positive staining. The cut off was used at 10%.
RESULTS
1. CpG sites
The Bisulfite sequencing method successfully revealed 32 CpG sites, the numbers started from 5’ end of the PCR products were CpG sites no. 1, 2 .... till 32 in sequence. From the mapping to the gene bank data, the primer used in the previous MSP study was composed of CpG sites no. 10, 11, 12, 13, 20, 21 and 22 (totally 7 sites).
2. Candidate active and inactive CpG sites
The candidate inactive CpG sites were derived from the group of hypermethylation with protein expression. The methylated sites in the two samples comprised sites no. 3, 7, 11, 12, 13, 14, 16, 17, 19, 21, 22, 23, 24, 25, 26 and 27 were not able to silencing the gene on encoding the protein (Table 2). Five of which, CpG sites no. 11, 12, 13, 21 and 22, were components of the primer of MSP.
The candidate active CpG sites were the rest of the CpG sites of the primer of MSP. They were CpG sites no. 10 and 20.
3. Verified active CpG sites
From Table 3, the samples of the group of MSP-related hypermethylation and non-protein expression revealed 6 out of 8 cases were found methylation involving either CpG no.10 or 20.
Table 2 Methylation status by Bisulfite sequencing technique in the group of MSP-related hypermethylation and protein expression
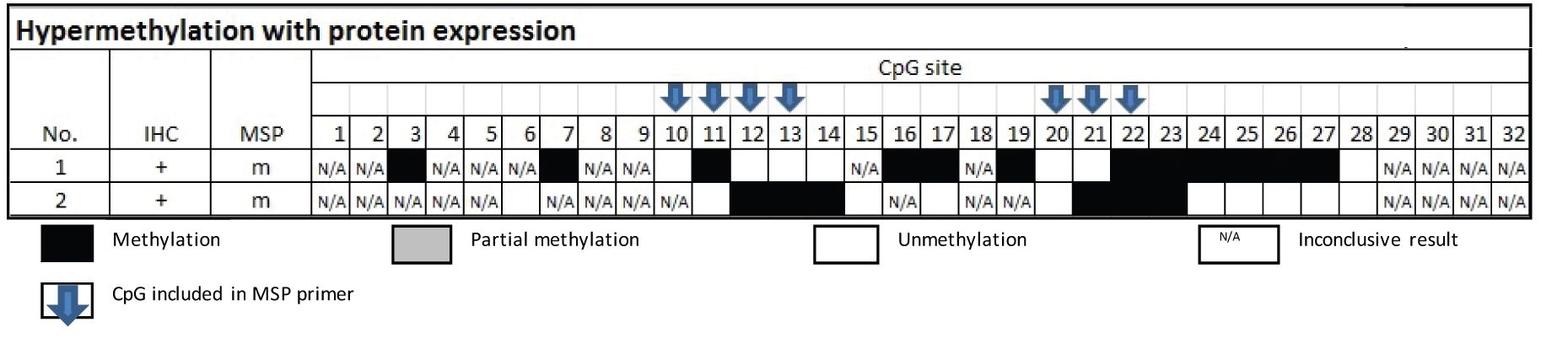
Table 3 Methylation status by Bisulfite sequencing technique in the group of MSP-related hypermethylation and non-protein expression
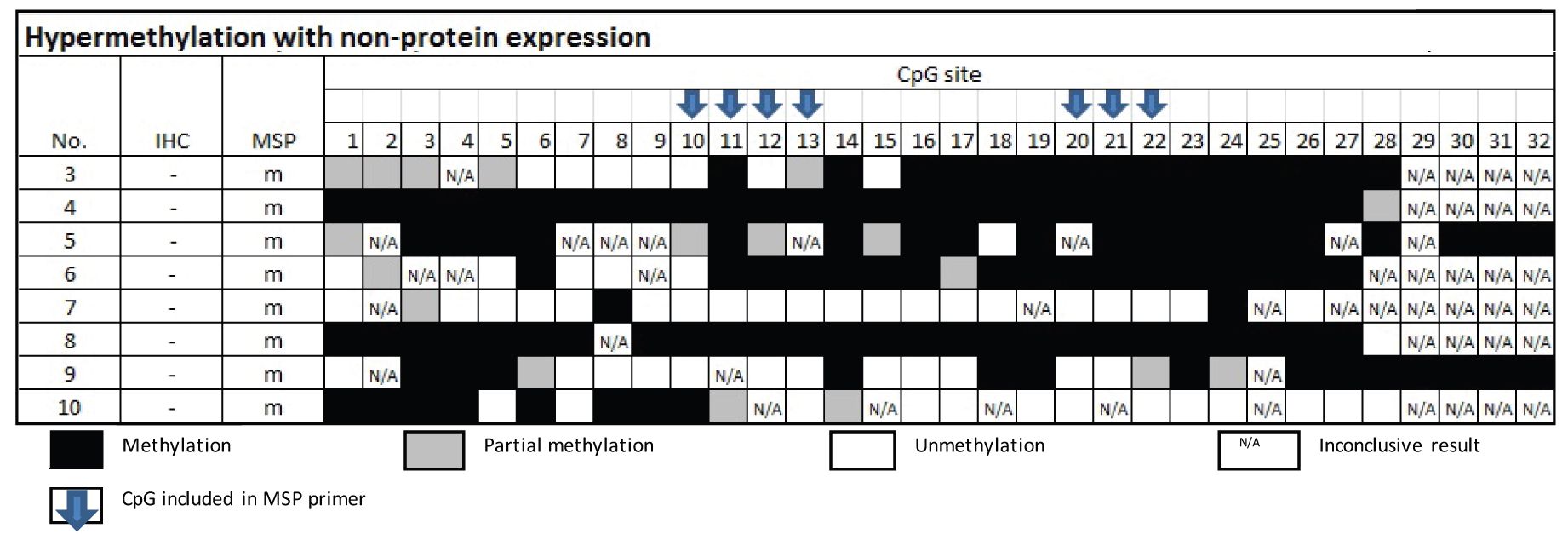
From Table 4, the samples of the group of MSP-related unmethylation with protein expression showed none of the 16 cases had methylation at CpG no. 10 and 20.
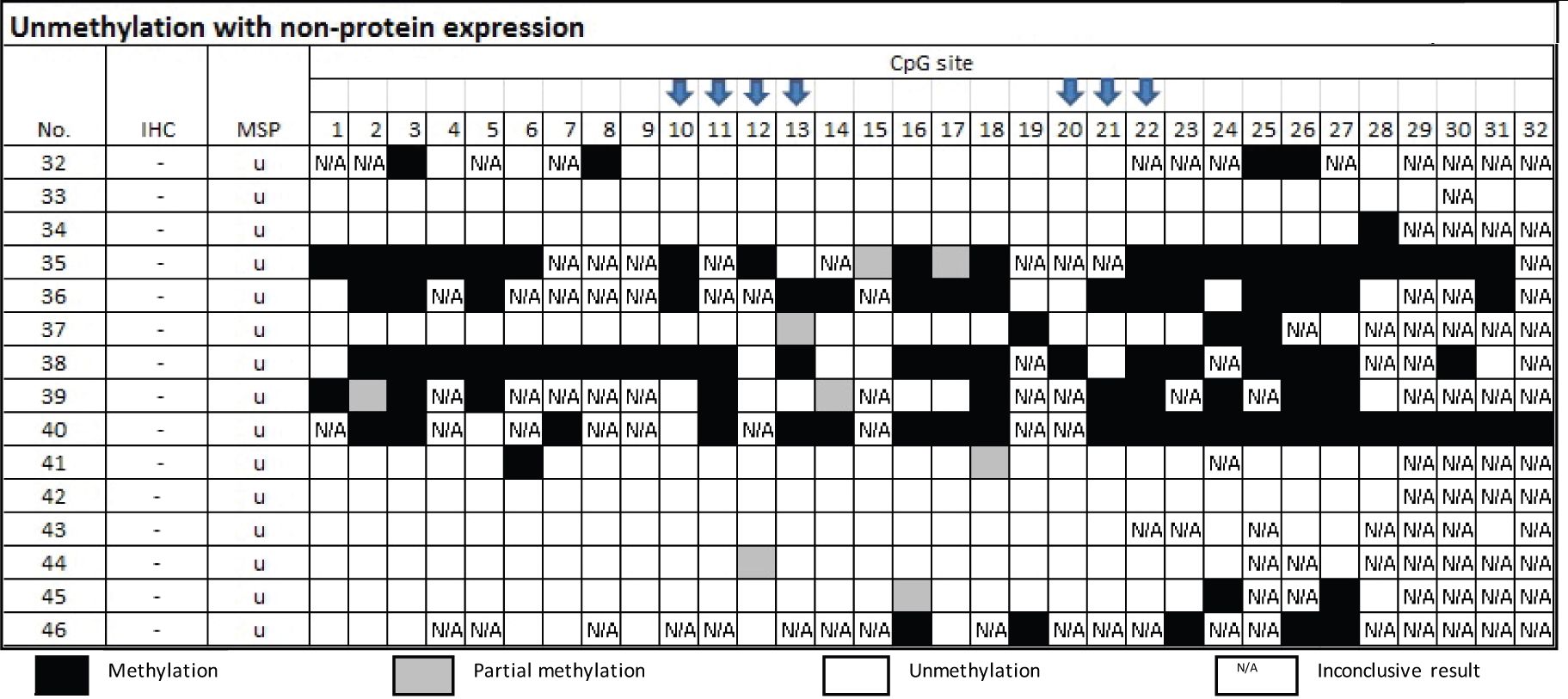
From Table 5, there were 15 samples of the group of MSP-related unmethylation with no protein, three cases of which were confirmed by bisulfite sequencing as truly unmethylation. Three cases out of the 12 cases that disclosed methylated cytosine by Bisulfite sequencing technique elicited either CpG 10 or 20.
DISCUSSION
DNA methylation is an epigenetic mechanism that is associated with many phenomena such as gene expression, genomic imprinting, and X-chromosome inactivation4. The mechanism on the protein expression is likely complicated; the attributed factors might include level and location of methylation2.
Table 4 Methylation status by Bisulfte sequencing technique in the group of MSP-related unmethylation
and protein expression
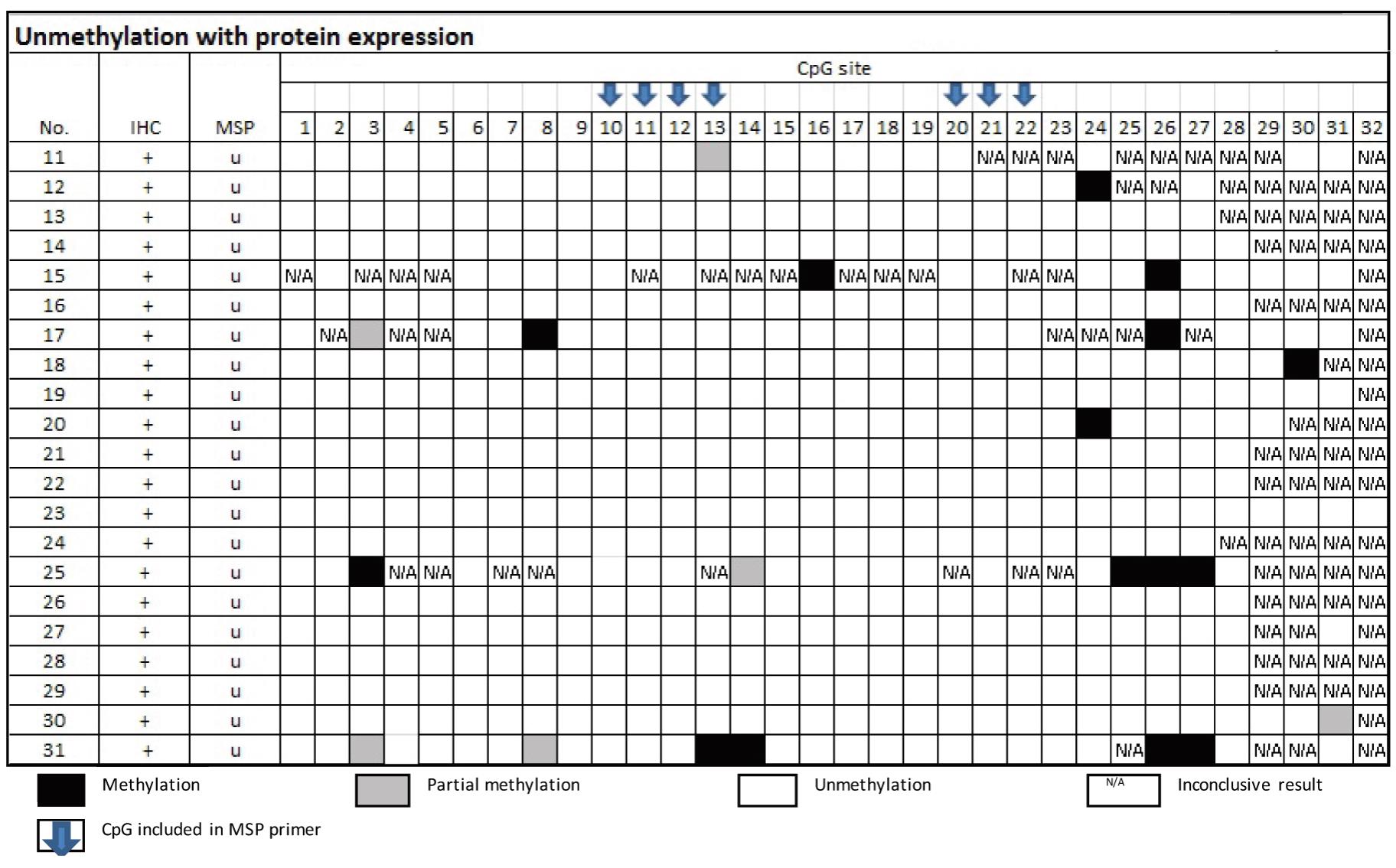
Table 5 Methylation status by Bisulfite sequencing technique in the group of MSP-related unmethylation and non-protein expression
DNA methylation is the adding of methyl group into 5-carbon position of cytosine that interferes the binding of transcription factor and thus protein expression56. As natural regulation, the level of methylation can be found on reducing or inhibiting the expression of consequent protein1,6. In pathologic process, DNA methylation is attributable to carcinogenesis as well as survival prognosis and treatment response1, 5 7 8 9 10. Jain S. et al used Bisulfite sequencing to study methylation status in individual CpG sites on promoter GSTP1 gene in various diseases of the liver and found that there are CpG site specific for disease subtypes and cancer7. Lin et al. reported the correlation between CpG island specific hypermethylation and disease severity of prostate cancer11. It is possible that some sites are active and some sites are not active. In addition, active sites may work in quantitative effects.
There were papers that quoted MSP positive and protein expression, though existed in a small percentage3,12,13,14. The previous study was shown by our group for the GSTP13 and there were for MGMT,
GSTP1 and ATM by other authors 12,13,14. These discordances between DNA methylation and the protein expression were probably from the MSP-based detections of some inactive methylated CpG sites. The individual positive sites could be only viewed with a DNA sequencing technique. In this study, we conducted with Bisulfite sequencing (BS) method to disclose the methylation in individual CpG sites. The MSP-based CpG sites are corresponding to the sites no. 10-13 and 20-22, totally 7 sites. We can find that the sites no. 10 and 20 are likely to be the active and all the other five CpG sites are inactive.
BS method is laborious , not suitable for use as a routine medical test. It is essential when individual CpG methylation status need known. In addition, the method is able to reveal a long segment of DNA region and to show whether the site is partially or fully methylated. In this study, we could demonstrate the CpG sites as far as 32 sites and with some partially methylated sites and much more sites with full methylation.
Although we experimented with a limited number of samples, the findings are adequate for the explanation; however repeated studies in a larger sample size is encouraging in order to achieve a detailed solution. From our findings, the methylated CpG sites that protein GSTP1 existed are regarded as inactive. There are 16 sites namely no. 3, 7, 11, 12, 13, 14, 16, 17, 19, 21, 22, 23, 24, 25, 26 and 27. The discovery of CpG no. 10 and 20 as active sites is not explicit since the other 14 sites that were not included in the candidate inactive group are await verification. The effect of the level of methylation is also not exhibited herewith since there was not any partial methylation of the CpG no.10 and 20 belonging to the protein presence group. The only partially methylated cytosine found at CpG no.10 was associated with fully methylated CpG sites outside the candidate inactive CpG sites.
MSP is the commonly used technique due to high sensitivity, specificity and l [3, 13, 15]. It is feasible to routine. Nevertheless, the reliability is dependent greatly on the primer design. We had tried our best to search the primer that had been used for the MSP to detect DNA methylation on the promoter of GSPT1 gene and found that all the published works quoted the primer from one single source that the specificity was unknown [16]. Based on our protein expression results, the new primer design to cover more active CpG sites may need. This can be accomplished if we know the impact of the remaining CpG sites including CpG sites no. 1,2,4,5,6,8,9,15,18,28-32. On the other hand, in our current experiment, many inconclusive results of CpG sites as well as the unble sequenced CpG sites that were the no. 33-38 were probably limitation of the BS. Pyrosequencing might be the technique of choice for further investigation [17].
In conclusion, the impact of location of CpG sites on the silencing of the protein expression of GSTP1 is present. There are at least two active sites and sixteen inactive sites out of the 32 CpG sites that were investigated. Further studies to design a new primer for the MSP are encouraging.
Disclosure: The authors declare no conflict of interest in this study.
REFERENCES
1. Jovanovic J, Ronneberg J, Tost J, and Kris-tensen V. The epigenetics of breast cancer.Mol Oncol.4(June 2010): 242-54.
2. van Eijk KR, de Jong S, Boks MP, Langeveld T, Colas F, Veldink JH, et al. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics 2012, 13:636.
3. Pakdeethai S, Fongchaiya V, Pontheerat T, Lampenkhae K, and Sampatanukul P. Relationship between promoter methylation and protein expression of glutathione S-transferase gene class P1 in breast cancer. Asian ArchPath. 8(October 2012):45-53.
4. Phillips T. The role of methylation in gene expression. Nature Education 1(2008):1.
5. Mutirangura A. Quantitative PCR analysis for methylation level of genome: clinical implications in cancer. Asian Biomed. 1(August 2007):121-8.
6. Melanie E. DNA Methylation in cancer: too much, but also too little. Oncogene. 21(August 2002):5400-13.
7. Jain S, Chen S, Chang KC, Lin YJ, Hu CT, Boldbaatar B, et al. Impact of the location of CpG methylation within the GSTP1 gene on its specificity as a DNA marker for hepatocellular carcinoma. PLoS ONE 7 (April 2012):e35789.
8. Gumy-Pause F, Pardo B, Khoshbeen-Boudal M, Ansari M, Gayet-Ageron A, Sappino AP, et al. GSTP1 Hypermethylation is associated with reduced protein expression, aggressive disease and prognosis in neuroblastoma. Gene Chromosome Canc. 51 (February 2012):174-85.
9. Arai T, Miyoshi Y, Kim SJ, Akazawa K, Maruyama N, Taguchi T, et al. Association of GSPT1 expression with resistance to docetaxel and paclitaxel in human breast cancers. Eur J Surg Oncol 2008; 34:734-8.
10. Gayatri S, Sameer M, and Rajinder P. Clinical significance of promoter hypermethylation of DNA repair in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci. 87(July 2010):83-91.
11. Lin PC, Giannopoulou EG, Park K, Mosquera JM, Sboner A, Tewari AK, et al. Epigenomic Alterations in Localized and Advanced Prostate Cancer. Neoplasia. 4(April 2013): 373-83.
12. Treilleux I, Chapot B, Goddard S, Pisani P, An-gele S, and Hall J. The molecular causes of low ATM protein expression in breast carcinoma; promoter methylation and levels of the catalytic subunit of DNA-dependent protein kinase. Histopathology. 51(July 2007):63-9.
13. Uno M, Oba-Shinjo SM, Camargo AA, Moura RP, Aguiar PH, Cabrera HN, et al. Correlation of MGMT promoter methylation status with gene and protein expression levels in glioblastoma. Clinics. 10(2011):1747-55.
14. Lee JS. GSTP1 promoter hypermethylation is an early event in breast carcinogenesis. Virchows Archiv. 450(June 2007):637-42.
15. Fraga MF. and Esteller M. DNA methylation: a profile of methods and application. Biotechnique. 33(September 2002):632-49.
16. Goessl C, Krause H, Muller M, Heicappell R, Schrader M, Sachsinger J, et al. Fluorescent Methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 60 (November 2000):5941-5.
17. Havik, AB., et al. MGMT promoter methyla-tion in gliomas assessment by pyrosequencing and quantitative Methylation-specific PCR. J Transl Med.6 (March 2012):10-36.


